Development of eScience methods for drug discovery
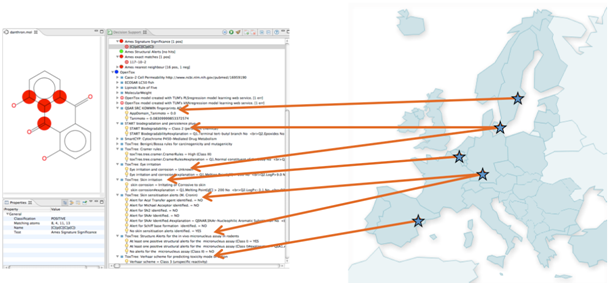
Our methods enable distributed predictions relevant for drug discovery, including predicting the toxicity for drug leads. The infrastructure requires an interoperable and standardized way of defining services and content over the Web, access to high-perofrmance computers, and is developed in collaboration with the EU FP7 project OpenTox (www.opentox.org). To the left we see a prediction in the Bioclipse workbench (www.bioclipse.net), which enables scientists to take advantage of the developed methods.
The pharmaceutical industry is struggling with bringing new drugs to the market and is currently undergoing big changes, with predictive sciences having high priority. The field however is not prepared for the transition to become data- and computationally intensive. There is a clear need for improved methods and tools in the area of modeling, managing big data, and user-friendly tools for accessing the relevant technologies.
Aims
In this project we aim at developing methods for modeling pharmaceutical problems with Big Data and high computational demands, and also develop methods for distributed data and predictive modeling in computational toxicology. We aim to enable researchers to take advantage of the offerings of eScience by adapting the Bioclipse workbench (www.bioclipse.net) to deliver e-Science technologies to scientists with low computational expertise. The project is propelled by industrial use cases and will be carried out in close collaboration with AstraZeneca R&D.
Methods
e-Science methods such as high-performance computing enables users to access computational resources on demand. An interesting technology is cloud computing, where more complex software and services can be provisioned using images and enable utilization of elastic computational resources (we will initially use Amazon EC2). Moreover, as data sets increases we will apply sparse data representation to accommodate for large modeling tasks in drug discovery. To enable distributed we will use and extend the OpenTox infrastructure, which is based on a European FP7 project that enables distributed and federated service provisioning and related data management specifications (API, ontologies) in computational toxicology.
Research group
PIs: Profs. Jarl Wikberg and Ola Spjuth
Dept of Pharmaceutical Biosciences, Uppsala University
Associate Professor Pierre Nugues, PhD.
Dept of Computer Science, LTH, Lund University
Valentin Georgiev,
Dept of Pharmaceutical Biosciences, Uppsala University
Jonathan Alvarsson,
Dept of Pharmaceutical Biosciences, Uppsala University
Behrooz Torabi,
Dept of Pharmaceutical Biosciences, Uppsala University
Links and references
Spjuth, O., Eklund, M., Ahlberg Helgee, E., Boyer, S., and Carlsson, L. Integrated decision support for assessing chemical liabilities. J Chem Inf Model 51, 8 (Aug 2011), 1840–7.
Spjuth, O., Helmus, T., Willighagen, E. L., Kuhn, S., Eklund, M., Wagener, J., Murray-Rust, P., Steinbeck, C., and Wikberg, J. E. S. Bioclipse: an open source workbench for chemo- and bioinformatics. J Chem Inf Model 51, 8 (Aug 2011), 1840–7.
The Bioclipse workbench: www.bioclipse.net
The OpenTox project: www.opentox.org