Translating long-read sequencing and metabolomics to clinical applications
The emergence of high-throughput molecular biology technologies has opened up for studying biological phenomena at increasingly detailed level. These technologies, such as biological sequencing, are evolving to supersede most of the current assays, providing comprehensive and accurate results with an in many cases faster process and more affordable price. However, their widespread uptake in clinical care for diagnostic, prognostic, and therapeutic use has yet to be realized; in many cases due to challenges in data management and –analysis, as well as lack of bioinformatics expertise at hospitals.
Long-read single molecule sequencing is a novel technology that enables sequencing of large genetic regions on the level of single DNA molecules in a single experiment. This not only improves sensitivity but also enables studying the effect of compound mutations which has previously not been possible. However, the data generated is large and requires new types of analysis pipelines and automated systems to enable the uptake in clinical care.
Another recent technology is mass spectrometry-based metabolomics, which has received a lot of attention in the search of new diagnostic markers, however there are significant challenges in data analysis and management in order to validate the technology and enable it to reach clinical routine applications.
Aims
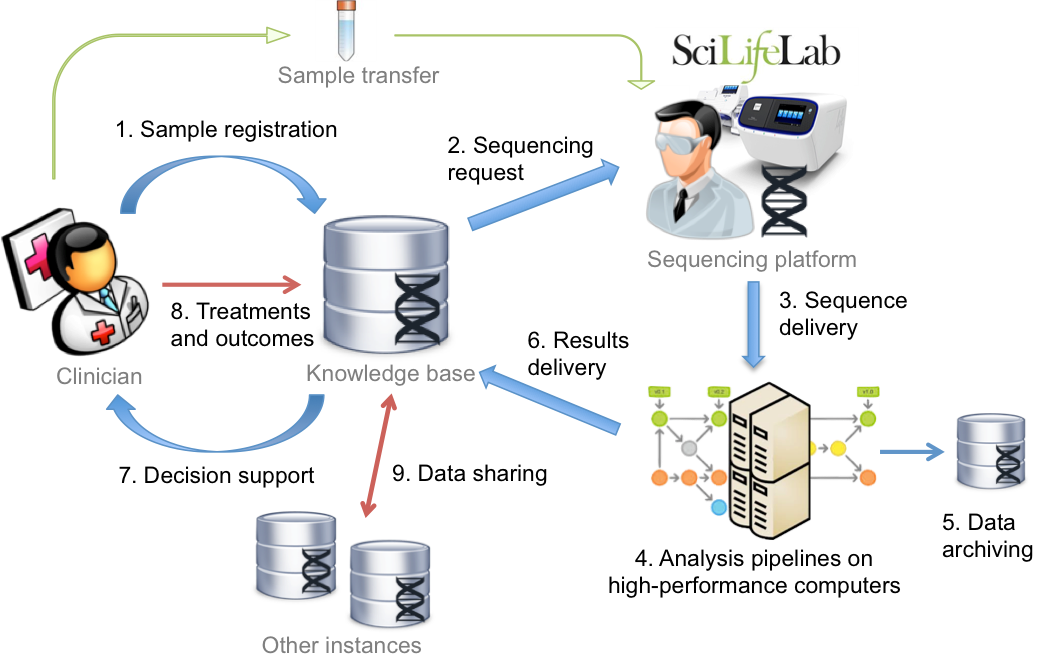
In this project, we seek to narrow the gap between academia and hospitals by developing methods and establishing systems to enable clinical care to take advantage of massively-parallel long-read single-molecule sequencing (LRSMS) and high-throughput metabolomics in routine diagnostics. Specific aims include the development of automated pipelines and data management systems for translating LRSMS for use in diagnosis of Chronic Myeloid Leukemia (CML). A second aim is to overcome the specific bioinformatics challenges when analyzing large-scale metabolomics data from neurological disorders, and pave the way towards ultimately bringing it into clinical routine practice.
The CML project is carried out in close collaboration with Clinical Genetics at Uppsala Academic Hospital (UAH), the National Genomics Infrastructure at Science for Life Laboratory (SciLifeLab), and is part of e-Science for Cancer Prevention and Control (eCPC) – a flagship project at the Swedish e-Science Research Center (SeRC). The metabolomics project is carried out in collaboration with the CARAMBA platform at Clinical Pharmacology, UAH and is also part of the EU H2020-project PhenoMeNal which has the aim to establish a European e-infrastructure and platform for data analysis in clinical metabolomics.
Methods
We develop automated data analysis pipelines that can run on high-performance and cloud computing resources to integrate and process large-scale data sets, and where results are delivered to structured databases. We also apply data mining and machine learning methods, and develop means for clinicians to interact and interpret the analysis results via web-interfaces. A necessity for this is scalable and interoperable e-infrastructures, and we use applied cloud computing and virtual infrastructures to deliver micro-service based architectures enabling researchers and clinicians to work with data on public and private Infrastructure-as-a-Service providers; the latter also facilitating operations on sensitive data.
Research group
PI:Assoc. Prof. Ola Spjuth
Dept of Pharmaceutical Biosciences and Science for Life Laboratory, Uppsala University
Wesley Schaal, Samuel Lampa and Marco Capuccini
Dept of Pharmaceutical Biosciences and Science for Life Laboratory, Uppsala University
Adam Ameur
National Genomics Infrastructure and Science for Life Laboratory, Uppsala University
Lucia Cavelier
Clinical Genetics Department, Uppsala Academic Hospital and Uppsala University
Kim Kultima and Stephanie Herman
Clinical Pharmacology Department, Uppsala Academic Hospital and the CARAMBA platform
Links and references
PhenoMeNal-H2020 website – https://phenomenal-h2020.eu
CARAMBA platform – www.caramba.clinic
Selected publications:
O. Spjuth, E. Bongcam-Rudloff, J. Dahlberg, M. Dahlö, A. Kallio, L. Pireddu, F. Vezzi, E. Korpelainen
Recommendations on e-Infrastructures for next-generation sequencing
GigaScience, vol. 5, no. 1, pp. 1-9, 2016.
O. Spjuth, J. Hastings, J. Dietrich, J. Heikkinän, N. Pedersen, J. Hottenga, S. Ripatti, P. Burton, I. Fortier, C. van Duijn, E. Wichmann, J. Rung, M. McCarthy, M. Allen, E. Raulo, I. Prokopenko, J. Karvanen, M. Perola, M. Kolz, E. J.C. de Geus, G. Willemsen, P. Magnusson, J-E. Litton, J. Palmgren, M. Krestyaninova, and J. Harris
Harmonising and linking biomedical and clinical data across disparate data archives to enable integrative cross-biobank research
European Journal of Human Genetics, 2016, 24, 521528
M. Dahlö, F. Haziza, A. Kallio, E. Korpelainen, E. Bongcam-Rudloff, and O. Spjuth
BioImg.org: A catalogue of virtual machine images for the life sciences
Bioinformatics and Biology Insights 2015:9 125-128
O. Spjuth, E. Bongcam-Rudloff, G. C. Hernandez, L. Forer, M. Giovacchini, R. V. Guimera, A. Kallio, E. Korpelainen, M. Kandula, M. Krachunov, D. P. Kreil, O. Kulev, P. P. Labaj, S. Lampa, L. Pireddu, S. Schönherr, A. Siretskiy, and D. Vassilev
Experiences with workflows for automating data-intensive bioinformatics
Biology Direct. 2015 Aug 19;10(1):43
A. Siretskiy, L. Pireddu, T. Sundqvist, and O. Spjuth
A quantitative assessment of the Hadoop framework for analyzing massively parallel DNA sequencing data
Gigascience. 2015 Jun 4; 4:26
S. Lampa, M. Dahlö, P. Olason, J. Hagberg, and O. Spjuth
Lessons learned from implementing a national infrastructure in Sweden for storage and analysis of next-generation sequencing data
GigaScience 2013, 2:9