Large-scale analysis of live cells
Obtaining quantitative data from large-scale experiments of live cells is of great importance for understanding molecular and cellular processes such as basic cellular processes in bacterial cells, and for design of personalized treatments based on cancer stem cells from patients. Live-cell experiments carried out using automated imaging systems often produce massive amounts of data containing far more information than can be digested by a human observer. A recurring task in many experiments is the tracking of large numbers of cells or particles and the analysis of their spatiotemporal behavior. The importance of using computer vision-based methods to accomplish this task is well recognized, but in practice investigators often encounter obstacles due to the lack of user-friendly software and infrastructure in terms computing resources and data storage.
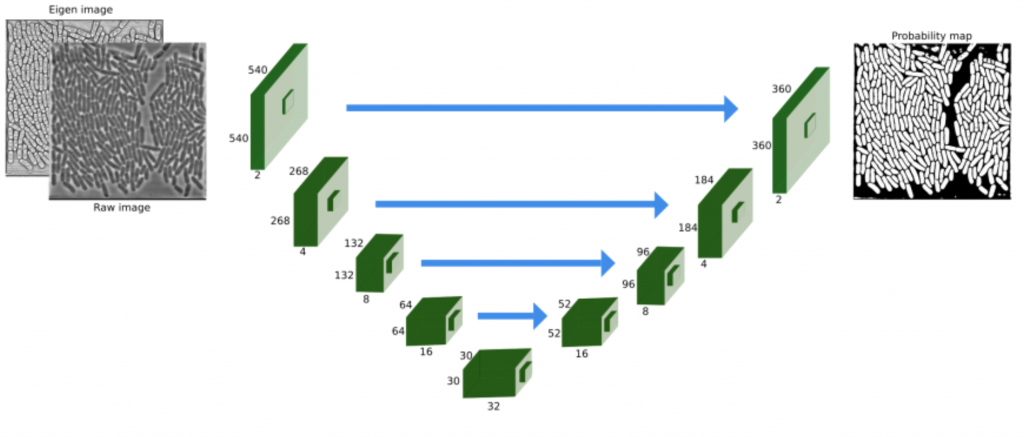
Aims
Optimizing software parameters for a new application usually requires knowledge of the underlying algorithms, making automated image processing far from routine. Instead, investigators often fall back to traditional manual data analysis, which is tedious, may bias data, and can limit an experiment’s value. However, manual input from an investigator is of high value for the optimization of automated approaches. The aim of this project is to maximize the use of the investigators’ biological knowledge to develop a flexible automated high-throughput tracking system that takes full advantage of computational resources and storage. The goal is to reduce time and algorithmic insight required by the investigator.
Methods
To improve parameter optimization in large-scale tracking experiments we combine existing and novel algorithms. We use the investigators’ visual/manual feedback as input for machine learning approaches to parameter optimization in an iterative process, with strong focus on image segmentation, which is a prerequisite for accurate object tracking. During the progress of the project, deep convolutional networks have proven to be very powerful for this type of tasks, and our research focuses on smart network designs, maximizing the use of manually curated training data, data augmentation, and pre-filtering techniques.
Research group
PI:Prof. Carolina Wählby
Div. of Visual Information and Interaction, Dept. Information Technology, Uppsala University
S.K. Sadanandan and O. Ishaq
Div. of Visual Information and Interaction, Dept. Information Technology, Uppsala University
Ö. Baltekin, A. Boucharin, and J. Elf
Dept. of Cell and Molecular Biology, Computational and Systems Biology, Uppsala University
K. E. G. Magnusson and J. Jaldén
ACCESS Linnaeus Centre, KTH Royal Institute of Technology
Links and references
Segmentation and track-analysis in time-lapse imaging of bacteria
IEEE Journal of Selected Topics in Signal Processing, October 15, 2015, 10(1):174-184
S.K. Sadanandan and C. Wählby
Feature Augmented Deep Neural Networks for Segmentation of Cells
Submitted, May 2016
O. Ishaq, S.K. Sadnandan, C, Wählby
Deep Fish: Deep Learning-based Classification of Zebrafish Deformation For High-throughput Screening
Submitted, June 2016.